import numpy as npimport pandas as pd
= 20 = 10 = n_cups - n_milk_first= np.concatenate((np.ones(n_milk_first),np.zeros(opposite)),axis= 0 ) #axis=0
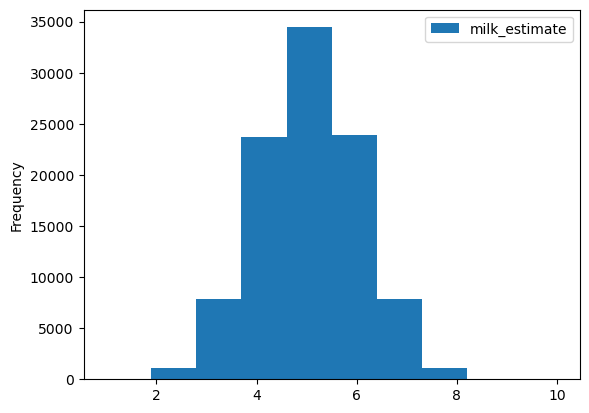
= 100000 = pd.DataFrame({"milk_estimate" :np.zeros(B)}) #딕셔너리를 데이터로 전달 for i in range (B):= np.random.choice(P,n_milk_first,replace= False )"milk_estimate" ] = np.sum (milk_first_choice)10 )
milk_estimate
0
6.0
1
6.0
2
4.0
3
5.0
4
6.0
5
5.0
6
4.0
7
5.0
8
4.0
9
5.0
<AxesSubplot:ylabel='Frequency'>
= 8 = milk_estimates[milk_estimates["milk_estimate" ] >= observed_val].shape
= 8 = 4 = n_cups - n_milk_first= np.concatenate((np.ones(n_milk_first),np.zeros(opposite)))= 4 = 1000000 = pd.DataFrame({"milk_estimates" :np.zeros(B)})for i in range (B):= np.random.choice(P,n_milk_first,replace= False )"milk_estimates" ] = np.sum (sample_milk)= estimates[estimates["milk_estimates" ]>= observed_val].shape= a/ Bprint (p_value)
= 100000 = 620 = np.random.binomial(1000 ,0.54 ,B)= np.sum (sample >= observed_val)/ B
import scipy.stats as stats= stats.binom(1000 ,0.54 )1 - binomial_dist.cdf(619 )
= "https://ilovedata.github.io/teaching/bigdata2/data/physical_test_2018_data.csv" = pd.read_csv(url)= physical_data[["TEST_AGE" , "TEST_SEX" , "ITEM_F001" , "ITEM_F002" ]].rename(columns = {"ITEM_F001" :"height" ,"ITEM_F002" :"weight" })10 )
TEST_AGE
TEST_SEX
height
weight
0
33
M
159.2
57.2
1
48
F
155.8
52.9
2
22
M
175.2
96.2
3
29
M
178.7
79.4
4
31
F
160.1
50.2
5
23
F
157.8
60.1
6
11
M
165.5
60.3
7
24
M
174.9
74.5
8
18
M
181.0
71.3
9
41
F
160.6
72.7
#randomnization = twogroup.shape= np.array([np.random.choice(["A" ,"B" ]) for i in np.arange(a)])"treatment" ] = treatment10 )
TEST_AGE
TEST_SEX
height
weight
treatment
0
33
M
159.2
57.2
A
1
48
F
155.8
52.9
B
2
22
M
175.2
96.2
B
3
29
M
178.7
79.4
A
4
31
F
160.1
50.2
B
5
23
F
157.8
60.1
B
6
11
M
165.5
60.3
A
7
24
M
174.9
74.5
B
8
18
M
181.0
71.3
A
9
41
F
160.6
72.7
B
= "https://ilovedata.github.io/teaching/bigdata2/data/drug.csv" = pd.read_csv(filename)
Placebo
Old
New
0
31
23
23
1
28
17
17
2
34
29
11
3
36
23
11
4
33
17
9
5
27
17
16
6
39
22
16
7
25
17
14
8
23
23
17
9
29
25
18
10
36
17
10
11
36
24
7
12
32
19
14
13
30
18
24
14
33
5
18
15
39
26
17
16
30
33
14
17
32
16
14
18
30
20
16
19
28
27
11
= three_drug_wide.melt(value_vars = ["Placebo" ,"Old" ,"New" ],value_name = "Value" ,var_name = "treatment" )
신약그룹이 기존약 그룹보다 효과가 있는가?
귀무가설 : 효과없다.
\[H_0 : \bar x_{old} - \bar x_{new} = 0 \]
데이터 관측 & 검정통계량 설정
\[\bar x_{old} - \bar x_{new} = 0 \]
def diff_mean(df,treatment):= df.loc[(df.treatment == treatment[0 ]) | (df.treatment == treatment[1 ])].reset_index(drop= True ) #reset_index 반드시 해주기 = twodrug.groupby("treatment" ).mean().reset_index()print (group_mean)= float (group_mean.loc[group_mean.treatment == treatment[0 ],"Value" ]) - float (group_mean.loc[group_mean.treatment == treatment[1 ],"Value" ])return test_stat"Old" ,"New" ])
treatment Value
0 New 14.85
1 Old 20.90
귀무가설 하에서의 분포 설정
= (three_drug_long.treatment == "Old" )| (three_drug_long.treatment == "New" )= three_drug_long[cond].reset_index(drop= True )= np.array(twodrug.treatment.sample(frac= 1.0 ,replace= False ))= twodrug"permuted_treat" ] = random_permuted_treat
treatment
Value
permuted_treat
0
Old
23
Old
1
Old
17
New
2
Old
29
Old
3
Old
23
New
4
Old
17
New
5
Old
17
Old
6
Old
22
Old
7
Old
17
Old
8
Old
23
Old
9
Old
25
Old
10
Old
17
New
11
Old
24
Old
12
Old
19
Old
13
Old
18
New
14
Old
5
Old
15
Old
26
New
16
Old
33
New
17
Old
16
New
18
Old
20
New
19
Old
27
New
20
New
23
New
21
New
17
New
22
New
11
New
23
New
11
Old
24
New
9
Old
25
New
16
New
26
New
16
Old
27
New
14
Old
28
New
17
Old
29
New
18
Old
30
New
10
Old
31
New
7
New
32
New
14
New
33
New
24
Old
34
New
18
Old
35
New
17
New
36
New
14
New
37
New
14
New
38
New
16
New
39
New
11
Old